Could a Server with 64 Cores be 100x Slower than my Laptop?
Yes
This blogpost is a rework of my old post from medium.com. You can find the original post here
A long time ago I asked on Twitter if someone could help me with a puzzling problem. A tool I was using utilized the scipy linear algebra package to perform the calculations. Most of the time was spent, running the pinv function, which makes calculates the inverse matrix. There are four functions in the scipy.linalg module, that can calculate the inverse matrix: pinv, pinv2, pinvh, and inv. The first three use different methods of calculating a pseudo-inverse matrix, and the last one explicitly calculates the inverse matrix, and they produce more or less the same result (within a rounding error in general). Tests on a local laptop have shown, that pinv is the slowest implementation, pinv2 goes next, then goes pinvh, and inv was the fastest. In fact, it was 15x faster, than pinv. This meant we could have made our code run 15x faster too if we replaced the pinv with inv in the tool’s source code!
Sidenote: Using inv is not recommended if you don’t know in advance, whether the inverse exists or not. But in our case, it almost always existed, so inv could be used as the default option, and we could still use pinv2 as a fallback.
Sidenote 2: If you are working with numpy, then be aware that numpy and scipy have different conventions on naming. numpy’s pinv is more or less equivalent to the scipy pinv2, so do not get confused with the naming in the tweet below.
Our code took 8 hours to finish on a real-world dataset. The opportunity to make it run in just 30 minutes was too good not to try! But then, something unexpected has happened. While inv was the fastest method locally, it was very slow on the server!
The timings of the rest 3 pseudo-inverse implementations were in the order that we have expected. But inv just didn’t comply with the expectations:
The wisdom of Twitter has offered me three ideas, all equally good:

Optimized compilation to the target is important, so Niko’s suggestion could be very real.

The memory layout of the arrays is also very important. If you have ever used numba to optimize your code (I have), you may have experienced, how much faster your code becomes, when numpy arrays are properly oriented in memory, so Moritz’s suggestion also seemed nice.
The last suggestion by Petar was, unfortunately, not preserved in the history. He suggested that an issue was with having more than one CPU. That was odd. I mean, it can’t possibly be, that a well-respected C-based BLAS package would be written in such a way that it would fail on servers, can it?
I think you can already guess, what has happened next.
Recompiling was absolutely useless. Memory layout had nothing to do with the case either. It turned out, that there are some concurrency issues with the inv. BLAS users can specify, how many threads BLAS can use. Here is an execution time for the inv applied to the test data, as a function of number of threads (notice, that y-axis is logarithmic):
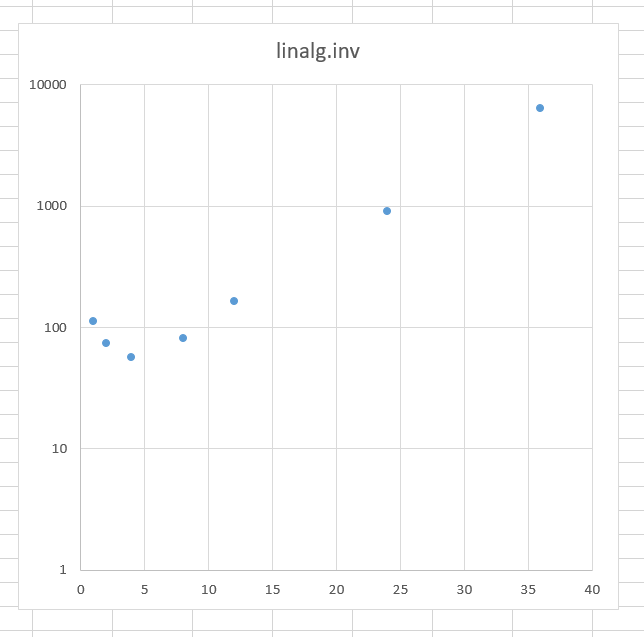
So, our solution was to limit the number of cores, to make the code run faster
Weirdly enough, this was not the case for all the mathematical functions! While inv had slowed down, np.dot behaved as expected. The execution time of np.dot becomes shorter when more cores are provided until it is saturated around 10–15 cores:
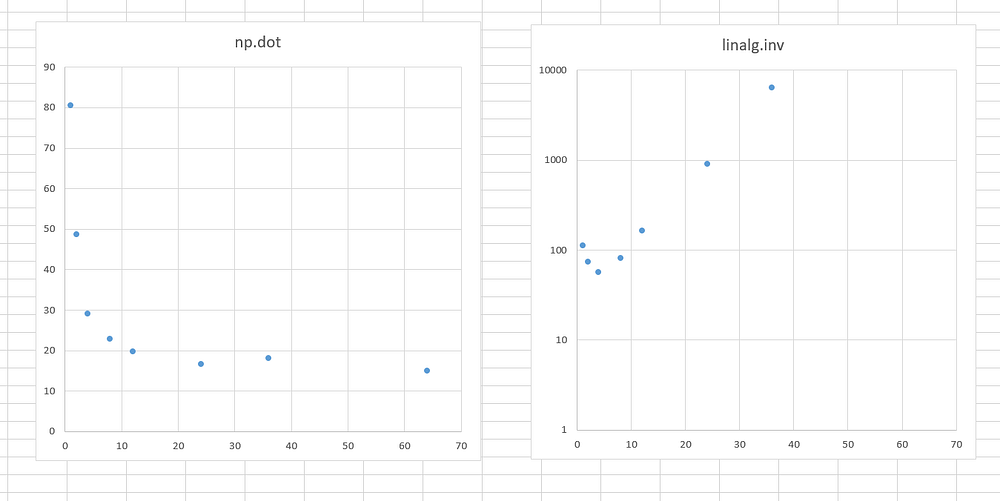
In the end, we have a limited number of cores to be used in our code to 6 as a compromise. The np.dot, which was the second most time-consuming function in our code, is fast enough when six cores are used. And for the scipy inv, this was the most optimal solution.
Did we achieve the 15x speed increase? Unfortunately, no. The pinv was not actually taking all the code execution time, and there were other parts, that were not affected by this change. But we still have shaved off a couple of hours of execution time, by literally replacing two characters in the code. If only it always was that easy.
Conclusions
- Measure the execution time using profiler for your code and code of the packages you are using.
- Try to find a single line or function, that takes at least 10–20% of the runtime.
- See, if you can speed it up.
- Prepare for unforeseen consequences, embrace them, and use them as an opportunity to learn something new.
- Always test your code in a production setup.